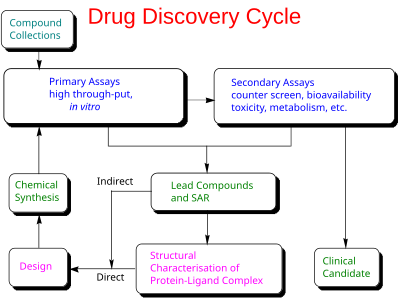
藥物探索
在醫學、生物技術和藥學領域,藥物探索是指一個新的候選藥物的發現過程。[1]
歷史上科學家通過鑑定傳統藥物中的活性成分或意外發現某物質具有療效從而發現藥物,如青黴素的發現。近代,科學家通過細胞或生物體篩選化合物庫中的合成小分子、天然產物或其提取物,旨在發現經典藥理學中具有理想療效的物質。人類基因組測序技術使得快速克隆和合成大量純化蛋白質成為可能。其中某些蛋白質靶標被認為與緩解或治療疾病有關,被稱為反向藥理學(Reverse pharmacology)。常規的藥物探索研究經歷如下幾個階段:先利用化合物庫中大量的化合物針對分離的蛋白質生物靶標進行高通量篩選(High throughput screening),然後將篩選出的活性化合物繼續在細胞中測試,最後將多輪測試中結果良好的化合物繼續進行動物實驗,以評價其治療活性或效力(Efficacy)。[2]
現代藥物探索包括以下幾個環節:苗頭化合物(Hit compound)的篩選與發現、 [3]藥物化學研究即從苗頭化合物至先導化合物(Hit to Lead)[4]和先導化合物最適化(Lead optimization)。其中最適化過程涵蓋了:增加與靶點的親和力(提高活性)、增加配體選擇性(減少副作用)、提高效力或活性、提高化合物代謝穩定性(以增加生物半衰期 )和提高藥物的口服生物利用度。只要找到某化合物可滿足以上所有條件,藥物開發過程就可以繼續推進,如若開發過程成功結束,將使用開發的新藥開展臨床試驗,以在人體上測試該新藥的安全性和有效性。[5]
現代新藥發現過程屬於資本密集型產業,這些資本涉及製藥企業的大量投資以及政府提供的政策補助和貸款擔保。儘管藥物科技和現代生物理論的不斷進步和對藥物探索頗有幫助,但迄今為止藥物探索仍然是一個漫長、昂貴、困難且低效的過程,發現全新療效的成功藥物的數量很少。[6]2010年統計每個新化學實體的研發費用約為18億美元。[7]21世紀針對藥物的基礎研究工作主要由政府和慈善組織資助,而藥物的後期開發主要由製藥公司或風投資本資助。[8]藥品的獲准上市前,必須經歷幾個臨床試驗階段,若成功才可通過新藥批准程式並上市銷售,在美國稱為新藥申請(New Drug Application,NDA)。
一個可取得商業或公共衛生領域成功的藥物探索過程,需要投資家、企業家、學術界、法律專利領域、政府監管領域和市場營銷之間的通力協作。[9]'罕見疾病患者人群較少,可預見其治療藥物無法取得商業上豐厚的回報,或在公共健康領域產生巨大影響。因此政府和眾多孤兒藥資助基金可幫助這些罕見病患者,讓其通過各種資助仍可獲得良好的藥物治療。[10][11][1][12]
歷史
編輯藥物在人體中的作用機制,是由藥物分子與生物大分子(通常為蛋白質或核酸)的特定相互作用介導的,基於以上觀點科學家得出論斷:藥品中具備的生物活性是由其中的單一化學物質產生的。從此藥用植物的粗品提取物被單一純淨的化學物質所替代成為標準藥物,這正是藥理學時代的開端。例如,鴉片是罌粟的粗品提取物,其具有鎮痛活性的單一化學成分是嗎啡[13];而洋地黃中的粗品提取物具有心臟興奮活性,其有效化學成分是地高辛[14][15]。使用有機化學研究天然產物中活性成分的學科稱為天然產物化學,[16][17]有機化學還促成了許多活性天然產物的全合成。[18][19][20]
歷史上,無論是粗品提取物還是純化學物質,均在不明生物靶標的情況下進行生物活性篩選工作。而先鑑定化學物質的結構,並用其驗證療效或相關生物活性,這種方法被稱為經典藥理學或正向藥理學[21],或稱表型藥物探索。[22]
後來,科學家針對已知的生理或病理途徑,設計且合成了一系列具相關活性的小分子,以避免對化合物庫篩選的依賴並取得了巨大的成功。例如:格特魯德·B·埃利恩(Gertrude Elion)和喬治·H·希欽斯在嘌呤代謝方面的工作,[23][24][25]詹姆士·W·布拉克在β受體阻滯劑和西咪替丁方面的工作,[26]以及遠藤章在他汀類藥物方面的工作。[27]另一個成功範例是大衛傑克(David Jack),他基於已知活性物質開發化合物類似物藥物,在葛蘭素史克製藥中參與或主導開發以下項目:第一個用於哮喘的甾體類吸入劑藥物;選擇性β2-腎上腺素能促效劑——雷尼替丁(西咪替丁的二代藥物),以及曲坦類藥物的開發工作。[28][29]
格特魯德·B·埃利恩與不到50人的小組一起開發嘌呤類似物,發現了首款抗病毒藥物和首個輔助人體器官移植的免疫抑制劑——硫唑嘌呤,並開發了首款緩解兒童白血病藥物。他一生參與了多個關鍵的腫瘤藥物、抗瘧疾藥物、抗菌藥物和痛風藥物的研發項目,並於1988年,獲頒諾貝爾生理學或醫學獎。[30][31][32]
靶標
編輯靶點或稱靶標(Target)在生物學中是指生物體內,能夠被其他物質(配體或藥物分子)識別或結合的結構。在藥學中,靶點是指研究的病理學相關的細胞或其分子結構,開發的藥物會與之作用並治療疾病。[8]然而,在不完全理解靶標資訊的情況下,可將靶標區分為新靶標或已知靶標,通常由新藥開發公司對靶標進行區分。[8]根據2011年統計,約有435種人類基因組物質被FDA定義為批准藥物的治療藥物靶標。[34]
「已知靶標」是指有大量歷史文獻的支持下,對該靶標如何在正常生理學中發揮作用,以及該靶標如何參與人類病理學均有良好的科學性理解。[2]但已知靶標不意味着藥物的作用機制已被完全了解,[2]其科學背景資訊已經過大量研究,尤其是靶標的功能性資訊。通常「新靶標」不是已知靶標,而是在藥物探索中已有大量科學數據和研究資料的靶標。在藥物探索工作中,主要研究的靶標類型為蛋白質,例如G蛋白偶聯受體(GPCR)和蛋白激酶。[35]
篩選和設計
編輯藥物探索通常從確定疾病的靶標開始,然後利用高通量篩選(HTS)和大型化合物庫進行篩選,找到能與靶標結合的化合物。例如,如果靶標是新的G蛋白偶聯受體,可通過以上方法篩選化合物抑制或激動該受體的活性(參見拮抗劑和促效劑);而如果靶標是一種蛋白激酶,則需要篩選化合物抑制該激酶的能力。[36]
高通量篩選的另一個作用是測試某化合物對目標靶標的特異性。藥物開發者期待找到一種藥物分子,其只會影響目標靶標,而不影響體內其他相關靶標。因此,可將某化合物進行多個靶標的篩選,以確認該化合物「命中」目標靶標的同時是否會干擾其他相關靶標,即所謂的脫靶效應,這個過程稱為靶標的交叉篩選。[36]交叉篩選的必要性在於:化合物可命中的靶點越多意味着特異性越差,在臨床實驗中表現出的脫靶毒性的可能性也越大。[36]
早期篩選中通常找不到完美的候選藥物,所以第一階段可排除一些無法繼續開發的化合物;若某化合物在幾乎所有篩選中均可命中不同靶標,該類化合物被藥物開發者定義為「泛分析干擾化合物」,在此階段該類化合物將從庫中被剔除。[37][38][39]在篩選期間,如果發現幾種化合物具有類似程度的生物活性,且這些化合物具有某些共同的化學基團或特徵,則基團或特徵可能屬於藥效團。基於藥效團的研究,藥物化學家嘗試分析化合物的構效關係(SAR)來改善先導化合物(Lead compound)的各項藥物特性:[40][41]
整個篩選過程需要生物實驗與化學結構改造經歷反覆且多次迭代,在此期間,新化學實體的特性在生物實驗的結果反饋中不斷進行最適化,並挑選數據良好的化合物繼續開展體外和體內的生物實驗測試,最後將體內外實驗均良好的候選化合物用於所選疾病的動物模型中,以測試該化合物的體內的有效性和安全性。[40]
與藥物吸收相關的物理化學性質包括:電離(pKa)、溶解度和滲透性等。其中滲透性可通過平行人工膜滲透性測定(Parallel artificial membrane permeability assay,PAMPA)或Caco-2細胞測定。[42][43]Caco-2、胃腸道(GIT)和血腦屏障(BBB)等測試具有高度相關性的優點,但PAMPA法與之相比藥物損耗低且價格低廉,因此作為早期化合物滲透性的篩選工具而被廣泛使用。[44]
一系列參數可用於評估化合物的成藥性或類藥性(Drug-Like property),[45]如Lipinski五原則。[46][47]這些參數中有些可通過模型預測估算,如用於估算親脂性的參數有:分配係數(cLogP) 、分子量(Mw)、極性表面積(TPSA)。以及只有通過實驗測試才可知的參數,如:效力(或效價)、酶促清除率的其他體外測試項目等。此外如:配體效率(LE)[48]和親脂效率(LiPE)[49][50]等參數也可參與評估類藥性質。[51]
高通量篩選雖常用於新藥發現,但並非成功的唯一途徑。藥物設計還可基於一些具備藥物特性的分子進行開發,此類分子可從自然界的動植物中提取的天然產物,也可將已上市的藥物進行改造,即開發所謂的Me-too藥物。其他還有一些方法,如虛擬高通量篩選。其中所謂的虛擬篩選,是使用計算機生成的蛋白質靶點模型與虛擬化合物庫中的化合物進行對接(Docking)操作,以選出可能存在活性的苗頭化合物(Hit compound)。[36]
另一種藥物探索方法稱為藥物從頭設計,先推測一類化合物可匹配目標酶的活性位點(Active site)。例如,利用虛擬篩選和計算機輔助藥物設計識別可與目標蛋白質靶點模型相互作用的新化學實體。[52][53]分子建模[54]和分子動力學模擬對於提高先導化合物的活性和成藥性均起到指導作用。[55][56][57]
藥物探索模式正發生轉變,從昂貴且覆蓋化學空間(結構多樣性)有限的化合物庫中進行高通量篩選,逐漸轉變為較小的化合物庫(不超過幾千種化合物)篩選。這些新的模式包括:基於片段的先導化合物發現(Fragment-based lead discovery, FBDD)[58][59][60][61]和蛋白質導向的動態組合化學(Protein-directed dynamic combinatorial chemistry)。[62][63][64][65][66]以上方法中使用的配體小分子通常體積較小,且與靶蛋白的結合親和力較HTS中篩選出的化合物更弱。首輪化合物被篩選出後,需要通過有機合成對小分子進行化學修飾,使之成為先導化合物。運用蛋白質X射線晶體學,可鑑定化合物與蛋白質片段形成複合物的三維空間結構,通過該結構可進一步指導先導化合物的修飾與最適化。[67][68][69]這些新模式的優點在於:篩選更有效率,且與HTS相比化合物庫雖然較小,但通常覆蓋的化學空間(Chemical space)很大。
表型篩選(Phenotypic screen)也是藥物探索新的起點之一。[70][71]多種動物或細胞模型可供篩選藥物,包括:酵母、斑馬魚、蠕蟲、永生細胞系、原代細胞系、患者來源的細胞系和完整動物模型。這些篩選旨在尋找可逆轉疾病表型的化合物,其表型包括:死亡、蛋白質聚集、突變蛋白質表達或細胞增殖。成功入選的化合物,繼續使用更全面的細胞模型或生物體進行篩選。此時當模型的價格或時間成本高昂時,可使用較小的篩選庫進行。[72]通常,表型篩選出的化合物雖然具有藥效,但其確切作用機制(MOA)卻是未知的,後續需要大量的目標逆卷積(Deconvolution)實驗來確定其作用機制。化學蛋白質組學(Chemoproteomics)的發展,為藥物設計提供了許多策略,以確定此類表型篩選藥物與靶標相互作用的機制。[73]
一旦確定先導化合物(Lead compound),即候選化合物具有足夠與靶標結合的活性和選擇性,也具有良好的類藥性。即可挑選其中的一至兩個系列化合物用於後續的藥物開發。其中最好的化合物被稱為先導化合物,另一個則被指定為備選化合物。[74][75][76]
源於天然產物的藥物
編輯傳統意義上,發現藥物和具有生物活性的化學物質,是基於研究有機體產生的化合物如何影響其他生物體活動的基礎之上。[78]
儘管組合化學(Combinatorial chemistry)已成為先導化合物發現中的關鍵一環,而天然產物仍然在藥物探索中發揮着重要作用。[79]2007年的一份報告報道了1981至2006幾年間,開發的974個小分子新化學實體中,63%為天然產物衍生物或天然產物的半合成衍生物。對於某些治療領域,例如抗菌藥物、抗腫瘤藥物、抗高血壓藥物和抗炎藥物,天然產物的比例更高。[80]天然產物是現代抗菌藥物開發中的新化學結構的重要來源之一。[81]
源於植物的藥物
編輯動植物產生的許多次級代謝產物具有潛在的藥用治療性質。這些次級代謝物包含以下活性:結合蛋白質或受體、酶等靶點並改變其生物功能。因此,動植物相關的天然產物常被用作藥物探索的起點。[82][83][84][85][3]
歷史
編輯西方直至文藝復興時期,絕大多數使用的西藥是植物提取物。[86]基於植物作為藥物探索的重要來源,逐漸形成了具有藥效潛力的植物資訊庫。[87]植物不同的部位(如根、葉和花)會產生不同的代謝物和激素,這些知識對於準確識別生物活性和植物藥理學屬性至關重要。[87][88]鑑於各個國家藥品監管機構的監管要求,鑑定天然產物並將其作為新藥推向市場會經歷嚴格的審批過程。[89]
茉莉酸
編輯茉莉酸(Jasmonate)會影響到細胞內信號的反饋機制。它們通過蛋白酶抑制劑誘導細胞凋亡[90][91]、蛋白質級聯(Protein cascade)[90]、具備防禦功能,[92]並調節植物對不同生物和非生物脅迫的反應。[92][93]茉莉酸還具有通過釋放代謝物誘導膜去極化,以直接作用於線粒體膜的能力。[94]
茉莉酸衍生物(JAD)對於植物細胞的傷口反應和組織再生非常重要。研究表明茉莉酸衍生物對人體表皮層具有抗衰老作用。[95]研究推測其與蛋白多糖(Proteoglycan,PG)和糖胺聚糖(Glycosaminoglycan,GAG)多糖相互發生作用,可幫助重塑細胞外基質的重要成分。[96]茉莉酸衍生物被發現在皮膚修複方面的研究,引發了科學家對於植物激素在藥物應用中的研究。[95]
水楊酸鹽
編輯水楊酸(Salicylic acid,SA)是一種植物激素,最初來源於柳樹皮,此後被發現在許多植物物種中存在。儘管科學家仍未完全了解水楊酸在植物中的作用,但可以確定其是植物免疫力的關鍵化學物質,[97]它參與了植物和動物組織的疾病和免疫反應。水楊酸可結合水楊酸結合蛋白(Salicylic acid binding proteins,SABP),已顯示其可對多種動物組織產生影響。[97]分離出的水楊酸化合物,首個發現的藥用屬性是治療疼痛和發燒。其次,它還在抑制細胞增殖中發揮積極作用,[90]並具有誘導白血病淋巴細胞凋亡及其他人類癌細胞凋亡的能力。[90]最常見的水楊酸類藥物是阿士匹靈,也稱為乙醯柳酸,其具有抗炎和解熱的作用。[97][98]
源於動物的藥物
編輯現代醫學中使用的一些藥物,其發現過程來源於動物體內,或是基於在動物發現的化合物。例如,抗凝藥物水蛭素,及其化學類似物比伐盧定(Bivalirudin)是基於一種(歐洲醫蛭)的唾液中的活性物質基礎上研發出的藥物。[99]用於治療二型糖尿病藥物艾塞那肽(Exenatide),是基於一種毒蜥(美國毒蜥)的唾液中分離的活性物質基礎上研發出的藥物。 [100]
源於微生物代謝物的藥物
編輯微生物為生存會爭奪空間和營養。為了適應環境存活,許多微生物會避免與其他微生物競爭,並演化出避免其他微生物增殖的能力,因此抗菌藥物的主要研發基礎就是微生物。鏈黴菌分離株(Streptomyces isolate)是非常有價值的抗生素來源,其被稱為藥用黴菌。另一個通過微生物的防禦機制而發現的抗生素的經典例子,是1928年科學家通過被青黴菌污染的細菌培養物中發現了青黴素。
源於海洋無脊椎動物的藥物
編輯也可從海洋世界中發現新的生物活性物質。[101]1950年代科學家從海洋無脊椎動物中發現的阿拉伯糖核苷,首次證明核糖和脫氧核糖以外的糖類化合物可衍生出具有生物活性的核苷類化合物。[102]直至2004年,首個海洋動物的衍生藥物才獲得批准,如錐形蝸牛毒素:齊考諾肽(Ziconotide)也稱為 Prialt,可治療嚴重的神經性疼痛。其他海洋動物衍生藥物目前正在開展對於包括:癌症、抗炎和疼痛等適應症的臨床試驗。[103]
化學多樣性
編輯組合化學可高效地生成大型篩選化合物庫,以滿足高通量篩選需求。然而經過二十年的組合化學的相關研究和實際運用,有藥學家指出:儘管組合化學基於高效合成提供了大量的化合物,但先導化合物或候選藥物的數量並未增加。[80]這驅使藥學家開始研究與現有藥物或天然產物相比,組合化學產出的化合物的結構差異性。
化學資訊學定義了化學多樣性(Chemical diversity)這個概念,其描述為化合物根據其物理化學特性在化學空間中的分佈情況,通常用化學多樣性評價組合化學庫與天然產物之間的差異。合成化合物庫或組合化合物庫只可涵蓋有限且單一的化學空間,而現有已上市的藥物尤其是天然產物,均表現出更強的化學多樣性,它們在化學空間中表現的更均勻且廣泛地分佈。[79]
組合化合物庫中與天然產物最顯著的區別在於手性中心的數量,一般天然產物中手性中心相較更多。其次天然產物比庫中的化合物的結構剛性更強,且在芳香結構的數量上更少。其他的化學差異還包括雜原子的性質如:天然產物中富含氧和氮元素,而合成化合物中更常見硫和鹵素元素;以及天然產物中的非芳香族不飽和鍵數量較高。由於結構剛性和手性均在藥物化學中被認為可提高化合物的特異性和藥物活性,因此有部分藥學家認為,使用天然產物作為潛在的先導化合物分子比組合化學庫相較更具優勢。[80]
篩選
編輯從天然產物中發現全新的具備生物活性的化學實體有兩種主要方法:
第一種被稱為天然物料的隨機收集和篩選。生物學尤其是植物學的知識,通常被用於識別有藥用價值的生物物種。這種方法的有效性建立在全球生物多樣性中只有一小部分曾經接受過藥物活性測試。此外,生物體能生活在物種豐富且複雜的環境中,勢必需要進化出防禦機制和競爭機制才能得以生存。藥物開發即可運用以上的機制挑選天然產物。[104]
源於動物、植物和微生物的複雜生態系統會產生新穎且具潛在生物活性的化合物,這些化合物值得在藥物開發過程中加以利用。成功運用這一策略的範例,是國家癌症研究所在1960年代開始的抗腫瘤藥物篩選項目。該項目找到了著名的抗腫瘤藥物:紫杉醇。其是從太平洋紫杉樹短葉紅豆杉中分離並鑑定出化學結構。[104]紫杉醇通過全新的抗腫瘤機制(穩定微管)顯示出其高活性,已獲准臨床用於治療肺癌、乳腺癌、卵巢癌以及卡波西肉瘤。[105][104][105]在21世紀初,藥物卡巴他賽(Cabazitaxel),一種由法國賽諾菲公司研製生產的藥物,其基於紫杉醇的衍生出的另一種抗腫瘤類似物,被證明對前列腺癌有效。該藥物同樣通過阻止微管的形成起藥效,體內的微管會在分裂細胞中拉開染色體。[106][107]
其他示例有:喜樹鹼類藥物(Camtothecin):拓撲替康(Topotecan) 、伊立替康(Irinotecan)、[108]魯比替康(Rubitecan)和倍羅替康(Belotecan)。 鬼臼毒素類藥物:依託泊苷(Etoposide)和替尼泊苷(Teniposide)[109][110]。蒽環類藥物:阿克拉黴素(Aclarubicin)、柔紅黴素(Daunorubicin)、多柔比星(Doxorubicin)、表柔比星(Epirubicin)、[111][112]伊達比星(Idarubicin)、氨柔比星(Amrubicin)、吡柔比星 (Pirarubicin)、戊柔比星(Valrubicin)、和佐柔比星(Zorubicin)。 以及蒽二酮類藥物:米托蒽醌(Mitoxantrone)和匹衫瓊(Pixantrone )。[113][114][115]
第二種方法主要源於民族植物學,即研究植物在社會中的普遍用途以及民族藥理學,民族藥理學是民族植物學內部的一個領域,專門研究醫學用途。[116][117]
青蒿素是一種源自青蒿植物開發的抗瘧藥,自公元前200年以來青蒿一直用於中藥,是一種用於聯合治療多重耐藥性惡性瘧原蟲的藥物。[118][119]
結構解析
編輯化學結構解析對於了解藥物化學結構,進行構效關係研究,以及避免重複的化學結構研究工作至關重要。藥物探索的關鍵即在於發現新結構,該化學結構能創造新的治療手段或解決老藥的安全性問題。質譜法是一種基於化合物離子化的質荷比,識別化合物分子量或碎片分子量,以推測化合物結構的方法。大多數化合物在自然界中通常以混合物形式存在,[120]因此液相色譜法-質譜聯用(LC-MS)通常用於解析混合物中的不同化合物組分的分子量和相關化合物資訊。[121]已知化合物的鑑定,可以通過質譜數據庫進行比對,數據庫還可用於推測未知化合物的結構。確定天然產物化學結構主要運用核磁共振波譜儀進行,核磁圖譜可顯示結構中單個氫原子、碳原子、氟原子、磷原子或氮原子的資訊,以及這些不同原子之間互相的連接關係,從而推測化合物的結構資訊。[122][123][124]
新藥申請
編輯當一種藥物在其整個研究過程中證明其在預計的治療用途中是安全且有效時,藥品研發公司可以向藥監部門提交申請,即所謂的新藥申請(New drug application,NDA)。[125][126]藥監部門會檢查並研究藥物研發企業遞交的NDA相關資料,根據該藥物在臨床前研究和人體臨床實驗中的安全性、藥物作用的特異性,藥物的劑量,藥物的有效性等資訊,做出是否批准候選藥物科通過該申請的決定。通過NDA的藥物即可商業化上市並銷售。[127]
參見
編輯參考文獻
編輯- ^ 1.0 1.1 The drug development process. US Food and Drug Administration. 4 January 2018 [18 December 2019]. (原始內容存檔於2020-02-22).
- ^ 2.0 2.1 2.2 The drug development process: Step 1: Discovery and development. US Food and Drug Administration. 4 January 2018 [18 December 2019]. (原始內容存檔於2023-04-03).
- ^ 3.0 3.1 Helleboid S, Haug C, Lamottke K, et al. The Identification of Naturally Occurring Neoruscogenin as a Bioavailable, Potent, and High-Affinity Agonist of the Nuclear Receptor RORα (NR1F1). Journal of Biomolecular Screening. 2014;19(3):399–406. https://doi.org/10.1177/1087057113497095.
- ^ Herrmann, A., Roesner, M., Werner, T. et al. Potent inhibition of HIV replication in primary human cells by novel synthetic polyketides inspired by Aureothin. Sci Rep 10, 1326 (2020). https://doi.org/10.1038/s41598-020-57843-9.
- ^ The drug development process: Step 3: Clinical research. US Food and Drug Administration. 4 January 2018 [18 December 2019]. (原始內容存檔於2020-06-20).
- ^ Anson D, Ma J, He JQ. Identifying Cardiotoxic Compounds. Genetic Engineering & Biotechnology News. TechNote 29 (9) (Mary Ann Liebert). 1 May 2009: 34–35 [25 July 2009]. ISSN 1935-472X. OCLC 77706455. (原始內容存檔於21 September 2012).
- ^ Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nature Reviews. Drug Discovery. March 2010, 9 (3): 203–14. PMID 20168317. S2CID 1299234. doi:10.1038/nrd3078 .
- ^ 8.0 8.1 8.2 Current Model for Financing Drug Development: From Concept Through Approval. Institute of Medicine (US), Forum on Drug Discovery, Development, and Translation, National Academies Press, Washington (DC). 2009 [2023-05-28]. (原始內容存檔於2023-04-08).
- ^ Warren J. Drug discovery: lessons from evolution. British Journal of Clinical Pharmacology. April 2011, 71 (4): 497–503. PMC 3080636 . PMID 21395642. doi:10.1111/j.1365-2125.2010.03854.x.
- ^ Hadjivasiliou, Andreas, Orphan Drug Report 2014 (PDF), EvaluatePharma, October 2014 [28 June 2015], (原始內容存檔 (PDF)於2015-10-10)
- ^ Rich Daly (5 September 2002). "House Offers Incentives For Development of 'Orphan' Drugs". Congressional Quarterly Daily Monitor
- ^ Hall, Anthony K; Carlson, Marilyn R. The current status of orphan drug development in Europe and the US. Intractable and Rare Diseases Research. 2014, 3 (1): 1–7. ISSN 2186-3644. PMC 4204542 . PMID 25343119. doi:10.5582/irdr.3.1.
- ^ Narcotic Drugs Estimated World Requirements for 2008, Statistics for 2006.. New York: United Nations Pubns. 2008: 77 [2016-05-26]. ISBN 9789210481199. (原始內容存檔於2019-06-04).
- ^ Cartwright, Anthony C. The British Pharmacopoeia, 1864 to 2014: Medicines, International Standards and the State. Routledge. 2016: 183 [2016-12-22]. ISBN 9781317039792. (原始內容存檔於2017-09-08) (英語).
- ^ Hollman, A. Drugs for atrial fibrillation. Digoxin comes from Digitalis lanata.. BMJ (Clinical research ed.). 1996-04-06, 312 (7035): 912. PMID 8611904.
- ^ Webster's Revised Unabridged Dictionary. Natural product. Free Online Dictionary and C. & G. Merriam Co. 1913 [2023-05-30]. (原始內容存檔於2023-04-28).
A chemical substance produced by a living organism; – a term used commonly in reference to chemical substances found in nature that have distinctive pharmacological effects. Such a substance is considered a natural product even if it can be prepared by total synthesis.
- ^ All natural. Nature Chemical Biology. July 2007, 3 (7): 351. PMID 17576412. doi:10.1038/nchembio0707-351 .
The simplest definition for a natural product is a small molecule that is produced by a biological source.
- ^ K. C. Nicolaou; D. Vourloumis; N. Winssinger and P. S. Baran. The Art and Science of Total Synthesis at the Dawn of the Twenty-First Century (PDF). Angewandte Chemie International Edition. 2000, 39 (1): 44–122 [2020-06-25]. PMID 10649349. doi:10.1002/(SICI)1521-3773(20000103)39:1<44::AID-ANIE44>3.0.CO;2-L. (原始內容 (reprint)存檔於2017-05-17).
- ^ Nicolaou, K. C. & Sorensen, E. J. 1996, Classics in Total Synthesis: Targets, Strategies, Methods, New York:John Wiley & Sons, ISBN 978-3-527-29231-8
- ^ Nicolaou, K. C. & Snyder, S. A., 2003, Classics in Total Synthesis II: More Targets, Strategies, Methods, New York:John Wiley & Sons, ISBN 978-3-527-30684-8
- ^ Takenaka T. Classical vs reverse pharmacology in drug discovery. BJU International. September 2001, 88 (Suppl 2): 7–10; discussion 49–50. PMID 11589663. doi:10.1111/j.1464-410X.2001.00112.x .
- ^ Lee JA, Uhlik MT, Moxham CM, Tomandl D, Sall DJ. Modern phenotypic drug discovery is a viable, neoclassic pharma strategy. Journal of Medicinal Chemistry. May 2012, 55 (10): 4527–38. PMID 22409666. doi:10.1021/jm201649s.
- ^ Nobel prize winners who contributed to Transplantation - GIN. GIN. 2018-01-27 [2018-11-12]. (原始內容存檔於2018-11-12) (美國英語).
- ^ Elion GB. The quest for a cure. Annual Review of Pharmacology and Toxicology. 1993, 33: 1–23. PMID 8494337. doi:10.1146/annurev.pa.33.040193.000245.
- ^ Elion GB. The purine path to chemotherapy. Nobel Lecture 1988.. [2023-05-28]. (原始內容存檔於2008-07-24).
- ^ Black J. Drugs from emasculated hormones: the principles of synoptic antagonism. Nobel Lecture 1988.. [28 February 2014]. (原始內容存檔於2017-10-06).
- ^ Endo A. The discovery of the statins and their development.. [28 February 2014]. (原始內容存檔於2023-01-31).
- ^ Sir David Jack. The Daily Telegraph. 17 November 2011 [25 November 2011]. (原始內容存檔於2011-11-25).
- ^ Sir David Jack. The Herald. 19 November 2011.
- ^ Holloway, M. (1991) Profile: Gertrude Belle Elion – The Satisfaction of Delayed Gratification, Scientific American 265(4), 40-44.
- ^ Watts G. Obituary: Sir David Jack. The Lancet. 2012, 379 (9811): 116. S2CID 54305535. doi:10.1016/S0140-6736(12)60053-1 .
- ^ Mary Ellen Avery. Gertrude Belle Elion. 23 January 1918 — 21 February 1999. Biographical Memoirs of Fellows of the Royal Society. 2008-12-12, 54: 161–168 [2018-04-02]. ISSN 0080-4606. doi:10.1098/rsbm.2007.0051. (原始內容存檔於2017-04-01) (英語).
- ^ Qubit Pharmaceuticals Accelerates Drug Discovery with Hybrid Quantum Computing. HPC Wire. November 30, 2022 [2023-05-28]. (原始內容存檔於2023-02-06).
- ^ Rask-Andersen M, Almén MS, Schiöth HB. Trends in the exploitation of novel drug targets. Nature Reviews. Drug Discovery. August 2011, 10 (8): 579–90. PMID 21804595. S2CID 3328752. doi:10.1038/nrd3478.
- ^ Jacobson KA. New paradigms in GPCR drug discovery. Biochemical Pharmacology. December 2015, 98 (4): 541–555. PMC 4967540 . PMID 26265138. doi:10.1016/j.bcp.2015.08.085.
- ^ 36.0 36.1 36.2 36.3 Chen, Ya; Kirchmair, Johannes. Cheminformatics in natural product‐based drug discovery. Molecular Informatics. 2020-09-06, 39 (12): 2000171. ISSN 1868-1743. PMC 7757247 . PMID 32725781. doi:10.1002/minf.202000171.
- ^ Baker M. Deceptive curcumin offers cautionary tale for chemists. Nature. 9 January 2017, 541 (7636): 144–145. Bibcode:2017Natur.541..144B. PMID 28079090. doi:10.1038/541144a .
- ^ Dahlin JL, Walters MA. The essential roles of chemistry in high-throughput screening triage. Future Medicinal Chemistry. July 2014, 6 (11): 1265–90. PMC 4465542 . PMID 25163000. doi:10.4155/fmc.14.60.
- ^ Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry. April 2010, 53 (7): 2719–40. CiteSeerX 10.1.1.394.9155 . PMID 20131845. doi:10.1021/jm901137j.
- ^ 40.0 40.1 Hughes, JP; Rees, S; Kalindjian, SB; Philpott, KL. Principles of early drug discovery. British Journal of Pharmacology. March 2011, 162 (6): 1239–1249. PMC 3058157 . PMID 21091654. doi:10.1111/j.1476-5381.2010.01127.x.
- ^ Schneider, Gisbert. Prediction of Drug-Like Properties. Landes Bioscience. 2013 [20 November 2017]. (原始內容存檔於2023-04-17) (英語).
- ^ Gouriet, F; Saby, L; Delaunay, E; Cammilleri, S; le Dolley, Y; Riberi, A; Casalta, JP; Habib, G; Raoult, D. Incidental diagnosis of colonic tumor by PET/CT in infectious endocarditis.. The Journal of infection. 2013-07, 67 (1): 88–90 [2019-12-17]. PMID 23523828. doi:10.1016/j.jinf.2013.03.004. (原始內容存檔於2019-12-17).
- ^ Artursson, P; Palm, K; Luthman, K. Caco-2 monolayers in experimental and theoretical predictions of drug transport.. Advanced drug delivery reviews. 2001-03-01, 46 (1-3): 27–43 [2019-12-17]. PMID 11259831. doi:10.1016/s0169-409x(00)00128-9. (原始內容存檔於2019-12-17).
- ^ Ottaviani, Giorgio; Martel, Sophie; Carrupt, Pierre-Alain. Parallel Artificial Membrane Permeability Assay: A New Membrane for the Fast Prediction of Passive Human Skin Permeability. Journal of Medicinal Chemistry. 2006, 49 (13): 3948–3954. ISSN 0022-2623. PMID 16789751. doi:10.1021/jm060230+.
- ^ Edward HK, Li D. Druglike Properties: Concepts Structure Design and Methods from ADME to Toxicity Optimization. August 2008. doi:10.1016/C2013-0-18378-X.
- ^ Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. March 2001, 46 (1–3): 3–26. PMID 11259830. doi:10.1016/S0169-409X(00)00129-0.
- ^ Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies. December 2004, 1 (4): 337–341. PMID 24981612. doi:10.1016/j.ddtec.2004.11.007.
- ^ Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discovery Today. May 2004, 9 (10): 430–1. PMID 15109945. doi:10.1016/S1359-6446(04)03069-7.
- ^ Ryckmans T, Edwards MP, Horne VA, Correia AM, Owen DR, Thompson LR, Tran I, Tutt MF, Young T. Rapid assessment of a novel series of selective CB(2) agonists using parallel synthesis protocols: A Lipophilic Efficiency (LipE) analysis. Bioorganic & Medicinal Chemistry Letters. August 2009, 19 (15): 4406–9. PMID 19500981. doi:10.1016/j.bmcl.2009.05.062.
- ^ Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nature Reviews. Drug Discovery. November 2007, 6 (11): 881–90. PMID 17971784. S2CID 205476574. doi:10.1038/nrd2445.
- ^ Tetko IV, Bruneau P, Mewes HW, Rohrer DC, Poda GI. Can we estimate the accuracy of ADME-Tox predictions? (PDF). Drug Discovery Today. August 2006, 11 (15–16): 700–707 [2023-05-02]. PMID 16846797. doi:10.1016/j.drudis.2006.06.013. (原始內容 (pre-print)存檔於2013-09-12).
- ^ Rester U. From virtuality to reality – Virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Current Opinion in Drug Discovery & Development. July 2008, 11 (4): 559–68. PMID 18600572.
- ^ Rollinger JM, Stuppner H, Langer T. Natural Compounds as Drugs Volume I. Progress in Drug Research 65. 2008: 211, 213–49. ISBN 978-3-7643-8098-4. PMC 7124045 . PMID 18084917. doi:10.1007/978-3-7643-8117-2_6.
- ^ Barcellos GB, Pauli I, Caceres RA, Timmers LF, Dias R, de Azevedo WF. Molecular modeling as a tool for drug discovery. Current Drug Targets. December 2008, 9 (12): 1084–91. PMID 19128219. doi:10.2174/138945008786949388.
- ^ Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biology. October 2011, 9: 71. PMC 3203851 . PMID 22035460. doi:10.1186/1741-7007-9-71.
- ^ Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. Journal of Computer-Aided Molecular Design. January 2012, 26 (1): 15–26. Bibcode:2012JCAMD..26...15B. PMC 3268975 . PMID 22183577. doi:10.1007/s10822-011-9517-y.
- ^ Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, Kmiecik S. Protein-peptide docking: opportunities and challenges. Drug Discovery Today. May 2018, 23 (8): 1530–1537. PMID 29733895. doi:10.1016/j.drudis.2018.05.006 .
- ^ Erlanson DA, McDowell RS, O'Brien T. Fragment-based drug discovery. Journal of Medicinal Chemistry. July 2004, 47 (14): 3463–82. PMID 15214773. doi:10.1021/jm040031v.
- ^ Folkers G, Jahnke W, Erlanson DA, Mannhold R, Kubinyi H. Fragment-based Approaches in Drug Discovery (Methods and Principles in Medicinal Chemistry). Weinheim: Wiley-VCH. 2006. ISBN 978-3-527-31291-7.
- ^ Erlanson DA. Introduction to fragment-based drug discovery. Topics in Current Chemistry 317. June 2011: 1–32. ISBN 978-3-642-27539-5. PMID 21695633. doi:10.1007/128_2011_180.
- ^ Edward Z, Michael S. Fragment-based drug discovery a practical approach. Wiley. 2008.
- ^ Greaney MF, Bhat VT. Dynamic combinatorial chemistry: in drug discovery, bioinorganic chemistry, and materials sciences. New Jersey: John Wiley & Sons. 2010: 43–82.
- ^ Huang R, Leung IK. Protein-directed dynamic combinatorial chemistry: a guide to protein ligand and inhibitor discovery. Molecules. July 2016, 21 (7): 910. PMC 6273345 . PMID 27438816. doi:10.3390/molecules21070910 .
- ^ Mondal M, Hirsch AK. Dynamic combinatorial chemistry: a tool to facilitate the identification of inhibitors for protein targets. Chemical Society Reviews. April 2015, 44 (8): 2455–88. PMID 25706945. doi:10.1039/c4cs00493k .
- ^ Herrmann A. Dynamic combinatorial/covalent chemistry: a tool to read, generate and modulate the bioactivity of compounds and compound mixtures. Chemical Society Reviews. March 2014, 43 (6): 1899–933. PMID 24296754. doi:10.1039/c3cs60336a.
- ^ Hochgürtel M, Lehn JM. Fragment-based approaches in drug discovery . Weinheim: Wiley-VCH. 2006: 341–364. ISBN 9783527312917.
- ^ Caliandro R, Belviso DB, Aresta BM, de Candia M, Altomare CD. Protein crystallography and fragment-based drug design. Future Medicinal Chemistry. June 2013, 5 (10): 1121–40. PMID 23795969. doi:10.4155/fmc.13.84.
- ^ Chilingaryan Z, Yin Z, Oakley AJ. Fragment-based screening by protein crystallography: successes and pitfalls. International Journal of Molecular Sciences. October 2012, 13 (10): 12857–79. PMC 3497300 . PMID 23202926. doi:10.3390/ijms131012857 .
- ^ Valade A, Urban D, Beau JM. Two galactosyltransferases' selection of different binders from the same uridine-based dynamic combinatorial library. Journal of Combinatorial Chemistry. Jan–Feb 2007, 9 (1): 1–4. PMID 17206823. doi:10.1021/cc060033w.
- ^ Zheng W, Thorne N, McKew JC. Phenotypic screens as a renewed approach for drug discovery. Drug Discovery Today. November 2013, 18 (21–22): 1067–1073. PMC 4531371 . PMID 23850704. doi:10.1016/j.drudis.2013.07.001.
- ^ Swinney DC, Anthony J. How were new medicines discovered?. Nature Reviews. Drug Discovery. June 2011, 10 (7): 507–519. PMID 21701501. S2CID 19171881. doi:10.1038/nrd3480.
- ^ Brown DG, Wobst HJ. Opportunities and Challenges in Phenotypic Screening for Neurodegenerative Disease Research. Journal of Medicinal Chemistry. March 2020, 63 (5): 1823–1840. PMID 31268707. S2CID 195798523. doi:10.1021/acs.jmedchem.9b00797.
- ^ Moellering RE, Cravatt BF. How chemoproteomics can enable drug discovery and development. Chemistry & Biology. January 2012, 19 (1): 11–22. PMC 3312051 . PMID 22284350. doi:10.1016/j.chembiol.2012.01.001.
- ^ Marshall SF, Burghaus R, Cosson V, Cheung SY, Chenel M, DellaPasqua O, et al. Good Practices in Model-Informed Drug Discovery and Development: Practice, Application, and Documentation. CPT. March 2016, 5 (3): 93–122. PMC 4809625 . PMID 27069774. doi:10.1002/psp4.12049 .
- ^ Marshall S, Madabushi R, Manolis E, Krudys K, Staab A, Dykstra K, Visser SA. Model-Informed Drug Discovery and Development: Current Industry Good Practice and Regulatory Expectations and Future Perspectives. CPT. February 2019, 8 (2): 87–96. PMC 6389350 . PMID 30411538. doi:10.1002/psp4.12372 .
- ^ Van Wijk RC. Model-Informed Drug Discovery and Development Strategy for the Rapid Development of Anti-Tuberculosis Drug Combinations. Applied Sciences. 2020, 10 (2376): 2376. doi:10.3390/app10072376 .
- ^ Balatti, Galo, E.; Barletta, Patricio, G.; Perez, Andres, D.; Giudicessi, Silvana, L.; Martínez‐Ceron, María, C., Inamuddin; Altalhi, Tariq; Cruz, Jorddy N.; Refat, Moamen Salah El‐Deen , 編, Machine Learning Approaches to Improve Prediction of Target‐Drug Interactions, Drug Design Using Machine Learning 1 (Wiley), 2022-10-10: 21–96 [2023-01-27], ISBN 978-1-394-16628-2, doi:10.1002/9781394167258.ch2, (原始內容存檔於2023-01-27) (英語)
- ^ Allelopathy: a physiological process with ecological implications, Springer: 1, 2006, ISBN 978-1-4020-4279-9
- ^ 79.0 79.1 Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. Journal of Chemical Information and Computer Sciences. 2003, 43 (1): 218–27. PMID 12546556. doi:10.1021/ci0200467.
- ^ 80.0 80.1 80.2 Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. Journal of Natural Products. March 2007, 70 (3): 461–77. PMID 17309302. doi:10.1021/np068054v.
- ^ von Nussbaum F, Brands M, Hinzen B, Weigand S, Häbich D. Antibacterial natural products in medicinal chemistry—exodus or revival?. Angewandte Chemie. August 2006, 45 (31): 5072–129. PMID 16881035. doi:10.1002/anie.200600350.
The handling of natural products is cumbersome, requiring nonstandardized workflows and extended timelines. Revisiting natural products with modern chemistry and target-finding tools from biology (reversed genomics) is one option for their revival.
- ^ Li JW, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier?. Science. July 2009, 325 (5937): 161–5. Bibcode:2009Sci...325..161L. PMID 19589993. S2CID 207777087. doi:10.1126/science.1168243.
With the current framework of HTS in major pharmaceutical industries and increasing government restrictions on drug approvals, it is possible that the number of new natural product–derived drugs could go to zero. However, this is likely to be temporary, as the potential for new discoveries in the longer term is enormous.
- ^ Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era (PDF). Nature Reviews. Drug Discovery. 2015, 14 (2): 111–29 [2023-05-28]. PMID 25614221. S2CID 12369182. doi:10.1038/nrd4510. hdl:10072/141449 . (原始內容存檔 (PDF)於2023-01-31).
Here, we review strategies for natural product screening that harness the recent technical advances that have reduced [technical barriers to screening natural products in high-throughput assays]. The growing appreciation of functional assays and phenotypic screens may further contribute to a revival of interest in natural products for drug discovery.
- ^ Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. Journal of Natural Products. 2016, 79 (3): 629–61. PMID 26852623. doi:10.1021/acs.jnatprod.5b01055 .
... the utilization of natural products and/or their novel structures, in order to discover and develop the final drug entity, is still alive and well. For example, in the area of cancer, over the time frame from around the 1940s to the end of 2014, of the 175 small molecules approved, 131, or 75%, are other than "S" (synthetic), with 85, or 49%, actually being either natural products or directly derived therefrom.
- ^ Torre BG, Albericio F. The Pharmaceutical Industry in 2016. An Analysis of FDA Drug Approvals from a Perspective of the Molecule Type. Molecules (Basel, Switzerland). 2017, 22 (3): 368. PMC 6155368 . PMID 28264468. doi:10.3390/molecules22030368 .
The outputs from 2016 indicate the so-called small molecules are losing ground against biologics, biomolecules, and other molecules inspired [by] natural products
- ^ Sutton D. The Great Naturalists. London: Thames & Hudson, with the Natural History Museum. 2007: 32–37. ISBN 978-0-500-25139-3.
- ^ 87.0 87.1 Ahn K. The worldwide trend of using botanical drugs and strategies for developing global drugs. BMB Reports. March 2017, 50 (3): 111–116. PMC 5422022 . PMID 27998396. doi:10.5483/BMBRep.2017.50.3.221.
- ^ Wink M. Modes of Action of Herbal Medicines and Plant Secondary Metabolites. Medicines. September 2015, 2 (3): 251–286. PMC 5456217 . PMID 28930211. doi:10.3390/medicines2030251 .
- ^ Oishi S, Kimura SI, Noguchi S, Kondo M, Kondo Y, Shimokawa Y, Iwao Y, Itai S. New scale-down methodology from commercial to lab scale to optimize plant-derived soft gel capsule formulations on a commercial scale. International Journal of Pharmaceutics. January 2018, 535 (1–2): 371–378. PMID 29154803. doi:10.1016/j.ijpharm.2017.11.029.
- ^ 90.0 90.1 90.2 90.3 Fingrut O, Flescher E. Plant stress hormones suppress the proliferation and induce apoptosis in human cancer cells. Leukemia. April 2002, 16 (4): 608–16. PMID 11960340. doi:10.1038/sj.leu.2402419 (英語).
- ^ Zhang M, Zhang MW, Zhang L, Zhang L. Methyl jasmonate and its potential in cancer therapy. Plant Signaling & Behavior. 24 July 2015, 10 (9): e1062199. PMC 4883903 . PMID 26208889. doi:10.1080/15592324.2015.1062199.
- ^ 92.0 92.1 Turner JG, Ellis C, Devoto A. The jasmonate signal pathway. The Plant Cell. 2002, 14 (Suppl): S153–64. PMC 151253 . PMID 12045275. doi:10.1105/tpc.000679.
- ^ Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, Kazi AM, Gucel S. Jasmonates: Multifunctional Roles in Stress Tolerance. Frontiers in Plant Science. 2016, 7: 813. PMC 4908892 . PMID 27379115. doi:10.3389/fpls.2016.00813 (英語).
- ^ Rotem R, Heyfets A, Fingrut O, Blickstein D, Shaklai M, Flescher E. Jasmonates: novel anticancer agents acting directly and selectively on human cancer cell mitochondria. Cancer Research. March 2005, 65 (5): 1984–93. PMID 15753398. doi:10.1158/0008-5472.CAN-04-3091 .
- ^ 95.0 95.1 Michelet JF, Olive C, Rieux E, Fagot D, Simonetti L, Galey JB, Dalko-Csiba M, Bernard BA, Pereira R. The anti-ageing potential of a new jasmonic acid derivative (LR2412): in vitro evaluation using reconstructed epidermis Episkin™. Experimental Dermatology. May 2012, 21 (5): 398–400. PMID 22509841. doi:10.1111/j.1600-0625.2012.01480.x .
- ^ Henriet E, Jäger S, Tran C, Bastien P, Michelet JF, Minondo AM, Formanek F, Dalko-Csiba M, Lortat-Jacob H, Breton L, Vivès RR. A jasmonic acid derivative improves skin healing and induces changes in proteoglycan expression and glycosaminoglycan structure. Biochimica et Biophysica Acta (BBA) - General Subjects. September 2017, 1861 (9): 2250–2260. PMID 28602514. doi:10.1016/j.bbagen.2017.06.006.
- ^ 97.0 97.1 97.2 Klessig DF, Tian M, Choi HW. Multiple Targets of Salicylic Acid and Its Derivatives in Plants and Animals. Frontiers in Immunology. 26 May 2016, 7: 206. PMC 4880560 . PMID 27303403. doi:10.3389/fimmu.2016.00206 .
- ^ Pierpoint WS. Advances in Botanical Research 20. 1994: 163–235. ISBN 978-0-12-809447-1. doi:10.1016/S0065-2296(08)60217-7.
- ^ Taylor T, Campbell CT, Kelly B. A Review of Bivalirudin for Pediatric and Adult Mechanical Circulatory Support. American Journal of Cardiovascular Drugs. July 2021, 21 (4): 395–409. PMC 7654565 . PMID 33174088. doi:10.1007/s40256-020-00450-w.
- ^ Shyangdan DS, Royle P, Clar C, Sharma P, Waugh N, Snaith A. Glucagon-like peptide analogues for type 2 diabetes mellitus. The Cochrane Database of Systematic Reviews. October 2011, 2011 (10): CD006423. PMC 6486297 . PMID 21975753. doi:10.1002/14651858.cd006423.pub2.
- ^ Faulkner DJ, Newman DJ, Cragg GM. Investigations of the marine flora and fauna of the Islands of Palau. Natural Product Reports. February 2004, 21 (1): 50–76 [2023-05-28]. PMID 15039835. doi:10.1039/b300664f. (原始內容存檔於2023-02-05).
- ^ Holtzapple, M.T. HEMICELLULOSES. 2003: 3060–3071. doi:10.1016/B0-12-227055-X/00589-7.
- ^ Kollár P, Rajchard J, Balounová Z, Pazourek J. Marine natural products: bryostatins in preclinical and clinical studies. Pharmaceutical Biology. February 2014, 52 (2): 237–42. PMID 24033119. doi:10.3109/13880209.2013.804100 .
- ^ 104.0 104.1 104.2 Fischer, Janos; Ganellin, C. Robin. Analogue-based Drug Discovery. John Wiley & Sons. 2006: 512. ISBN 9783527607495. (原始內容存檔於2016-12-21) (英語).
- ^ 105.0 105.1 Taxol® (NSC 125973). National Cancer Institute:. [14 February 2016]. (原始內容存檔於5 September 2015). Wayback machine
- ^ Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nature Reviews. Cancer. April 2004, 4 (4): 253–65. PMID 15057285. S2CID 10228718. doi:10.1038/nrc1317.
- ^ Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK, et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Research. September 2011, 71 (18): 6019–29. PMC 3354631 . PMID 21799031. doi:10.1158/0008-5472.CAN-11-1417.
- ^ Nirmala, M. Joyce, A. Samundeeswari, and P. Deepa Sankar. 2011. "Natural Plant Resources in Anti-Cancer Therapy-A Review." Research in Plant Biology 1 (3): 1–14.. [2023-06-01]. (原始內容存檔於2018-10-19).
- ^ Osheroff Neil, Eukaryotic Topoisomerase II: characterisation of enzyme turnover, 1986, The Journal of Biological CHemistry, vol. 261, no. 21, pp. 9944-9950
- ^ Gordaliza M, García PA, del Corral JM, Castro MA, Gómez-Zurita MA. Podophyllotoxin: distribution, sources, applications and new cytotoxic derivatives. Toxicon. 2004, 44 (4): 441–59. PMID 15302526. doi:10.1016/j.toxicon.2004.05.008.
- ^ daunorubicin hydrochloride. The American Society of Health-System Pharmacists. [8 December 2016]. (原始內容存檔於8 January 2017).
- ^ Machida, Kodai; Masutan, Mamiko; Imatak, Hiroaki. Protein Synthesis in vitro: Cell-Free Systems Derived from Human Cells. Cell-Free Protein Synthesis. InTech. 2012-10-10. ISBN 978-953-51-0803-0.
- ^ Katzung, Bertram G. Cancer Chemotherapy. Basic and clinical pharmacology 10th. New York: McGraw-Hill Medical Publishing Division. 2006. ISBN 0-07-145153-6. OCLC 157011367.
- ^ Mazerski J, Martelli S, Borowski E. The geometry of intercalation complex of antitumor mitoxantrone and ametantrone with DNA: molecular dynamics simulations. Acta Biochim. Pol. 1998, 45 (1): 1–11. PMID 9701490.
- ^ The Random House Dictionary of the English Language Unabridged Edition, Jess Stein ed. in chief, Random House, New York 1966 p. 489
- ^ Acharya, Deepak and Shrivastava Anshu (2008): Indigenous Herbal Medicines: Tribal Formulations and Traditional Herbal Practices, Aavishkar Publishers Distributor, Jaipur, India. ISBN 978-81-7910-252-7. pp 440.
- ^ Acharya, Deepak and Shrivastava Anshu: Indigenous Herbal Medicines: Tribal Formulations and Traditional Herbal Practices. Aavishkar Publishers Distributor, Jaipur / India 2008, ISBN 978-81-7910-252-7, p. 440.
- ^ 米勒·路易斯(Louis H. Miller)和蘇新專(Xin-zhuan Su). 青蒿素:源自中草药园的发现. 《細胞》 (CAMBRIDGE, MA 02139, USA: Cell Press). 2011-09-16, 146 (6): 855–858 [2011-09-26]. ISSN 0092-8674. PMID 21907397. doi:10.1016/j.cell.2011.08.024. (原始內容存檔於2011-09-24).
- ^ 2015 诺贝尔生理学或医学奖:青蒿素与疟疾. [6 October 2015]. (原始內容存檔於2015-11-10).
- ^ Sparkman, O. David. Mass spectrometry desk reference. Pittsburgh: Global View Pub. 2000. ISBN 0-9660813-2-3.
- ^ Pitt, James J. Principles and Applications of Liquid Chromatography-Mass Spectrometry in Clinical Biochemistry. Clin Biochem Rev. February 2009, 30 (1): 19–34. PMC 2643089 . PMID 19224008.
- ^ Chaimbault, Patrick. The Modern Art of Identification of Natural Substances in Whole Plants. Jacob, Claus; Kirsch, Gilbert; Slusarenko, Alan; Winyard, Paul G.; Burkholz, Torsten (編). Recent Advances in Redox Active Plant and Microbial Products. Springer Netherlands. 2014-01-01: 31–94. ISBN 9789401789523. doi:10.1007/978-94-017-8953-0_3 (英語).
- ^ Background and Theory Page of Nuclear Magnetic Resonance Facility. Mark Wainwright Analytical Centre - University of Southern Wales Sydney. 9 December 2011 [9 February 2014]. (原始內容存檔於27 January 2014).
- ^ Shah, N; Sattar, A; Benanti, M; Hollander, S; Cheuck, L. Magnetic resonance spectroscopy as an imaging tool for cancer: a review of the literature.. The Journal of the American Osteopathic Association. January 2006, 106 (1): 23–27. PMID 16428685. (原始內容存檔於2013-04-07).
- ^ The Drug Development Process. U.S. Food and Drug Administration. 4 January 2018 [1 May 2018]. (原始內容存檔於2019-04-23).
- ^ The Drug Development Process. Step 4: FDA Drug Review. U.S. Food and Drug Administration. 4 January 2018 [1 May 2018]. (原始內容存檔於2019-04-22).
- ^ The drug development process. Step 4: FDA drug review. US Food and Drug Administration. 4 January 2018 [18 December 2019]. (原始內容存檔於2023-03-31).
延伸閱讀
編輯- Gad SC. Drug Discovery Handbook. Hoboken, N.J: Wiley-Interscience/J. Wiley. 2005. ISBN 978-0-471-21384-0.
- Madsen U, Krogsgaard-Larsen P, Liljefors T. Textbook of Drug Design and Discovery. Washington, DC: Taylor & Francis. 2002. ISBN 978-0-415-28288-8.
- Rasmussen N. Gene Jockeys: Life Science and the rise of Biotech Enterprise. Baltimore: Johns Hopkins University Press. 2014. ISBN 978-1-42141-340-2.